Biology Index
Where are we going with this? The information on this page should increase understanding related to this standard: Identify patterns of inheritance to predict genotype/phenotype and solve punnett square problems.
Article includes ideas, images, and content from Troy Smigielski (2022-01)
Law of Independent Assortment
and Non-Mendelian Genetics
(Onward and upward?)
So in previous articles, we explored genetics, many concepts of which grew out of Gregor Mendel's research.
Beyond the importance of how traits pass, he discovered something else.
Gregor Mendel also found out that genes are distributed to gametes independently from one another. He called this Law of Independent Assortment. This means that the version passed on of Gene A has no effect on the probability of getting either version of Gene B.
For example, just because a person has brown hair does not mean he or she will definitely have green eyes. Nor does it mean that the eye color will definately be green. Hair color does not determine eye color.
This is because the hair color gene is distributed (assorted) independently from the eye color gene. The gene for any one trait can be passed on with any combination of genes for other traits.
The Law of Independent Assortment greatly increases genetic variation because it allows for any possible combination of available genes. Combined with the crossover process in meiosis, independent assortment results in the vast range of trait variations genetic diversity that can be observed.
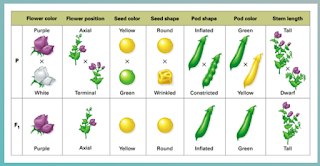
Mendel came to the understanding of independent assortment while doing his famous experiment with pea plants.
In additions to the color of the flower bloom, pea plants exhibit many other traits (including those pictured above). The dominant variations of the trait are at the top.
So, crossing the purebred (homozygous) varieties resulted in heterozygous plants in the F1 generation. When Mendel crossed those heterozygous plants, he ended up with a lot of variations in the F2 generation.
If he looked at any one trait, in the F2 generation, the predicted 1:2:1 genotype and 3:1 phenotype was observed. But many of the other traits would be mixed in.
This is because genes for the various traits are inherited separately from each other (Law of Independent Assortment).
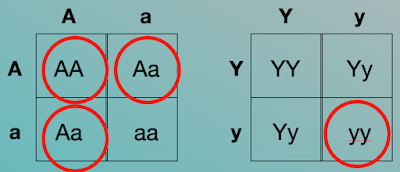
So, would it be possible for a cross between AaYy (axial; yellow) x AaYy (axial; yellow) to produce an axial, green-seed plant?
In order to have an axial plant, there must be at least one dominant "A" gene (AA or Aa). To have green seeds, requires that both of the genes be recessive (yy).
REMINDER: Each parent will contribute one allele for each gene.
Yes; it is possible because you could inherit an “A” from either parent resulting in the genotype “AA” (axial).
You could also inherit a “y” from both parents resulting in the genotype “yy” (green).
This is possible because these genes are inherited separately from each other (Law of Independent Assortment).
We can visualize that with two punnett squares. Because the genes assort independently, you can do two independent Punnett squares to find out if this is possible.
What if you wanted to examine two traits at the same time using a single Punnett square? Is that possible?
Dihybrid Cross Punnett Squares
It is possible to set up two or more traits in one Punnett square. There's a process…
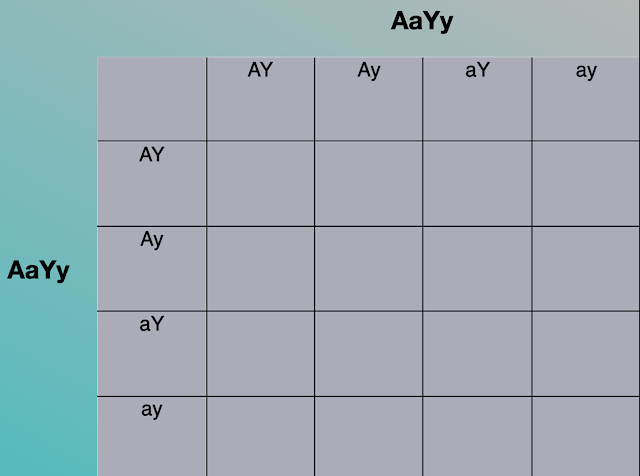
Recall that to look at one trait, the genotype of each parent is written, one at the top horizontally and one at the side vertically. (See above examples.)
When looking at more than one trait, the genotype will take on a form like…
Now, the same process is used… but with an added level of complexity.
To set up the square, write out the genotype for both, one at the top and one along the side.
In the above illustration, we have two homozygous parents:
AaYy
Number the genes 1 to 4…
1 2 3 4
A a Y y
Going along the top, you will put in the genes in this order:
Do the same thing down the side to get what is shown in the image above.
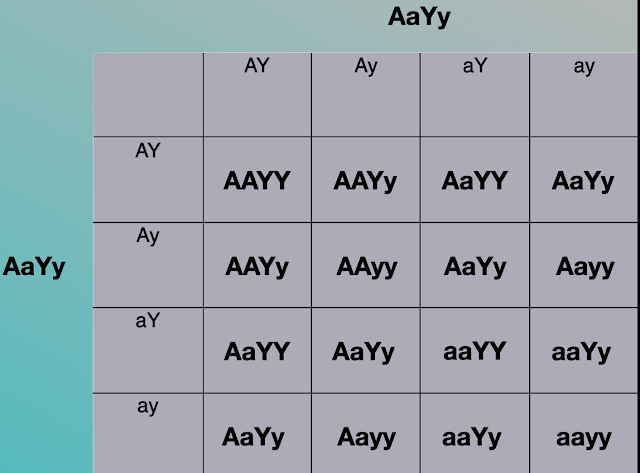
Then, you fill in each box much like with a one trait Punnett square. Take the genes from the top and side and order them into the boxes. You would end up with this:
A dihybrid Punnett square is an effective way to examine the offspring with consideration to two trait at the same time.
Law of Independent Assortment Recap
The Law of Segregation states that alleles separate independently during gamete formation.
Law of Independent Assortment states that genes separate independently during gamete formation.
Non-Mendelian Inheritance
Not all patterns of inheritance follow Mendelian Laws of dominance. There are other other systems by which traits can be passed from the parents to the offspring.
Incomplete dominance is where one allele does not show complete dominance over the other; they meet in the middle. Instead, of seeing the dominant
trait, the
traits of each allele can be thought to blend.
The most common example of this if crossing a red flower with a white flower and all offspring have pink flowers.
Though the homozygous parents are red and white, the heterozygous offspring are neither. They are a blend of the two.
Codominance is where both alleles appear in the phenotype; they are both expressed. A common example of this is blood type.
If blood type can be homozygous A or homozygous B, the heterozygous version of this is AB. Both traits are present.
You can think of incomplete dominance as blending of one version of the trait with the other. Red wasn't quite dominant enough to fully show up… it partly showed up as a blend. Dominance was incomplete.
For codominance, think of a team with co-captains or a club with co-presidents. You would say Bob and Mary are the co-chairs of our club. Codominance means that both of the versions of the trait show up. The flower is red and white, both. Co = and.
Polygenic traits are traits (physical appearance) that are affected by more than one gene. In actuality, most traits are polygenic. Common examples include height, skin color, eye color, and hair color.
To explain a little more, a trait like height might be made up of… let's say four genes. Each gene would appear as two alleles (versions) of the trait. Height would be the result of all of the genes combined.
Suppose someone's genotype for height looked like…
AaBBccDd
Those four genes would work together to establish the genetic potential for height. Of course, environmental factors such as nutrition would further influence height.
Someone with AAbbCcdd would have a different genetic potential for height.
Someone with aaBbccDD would also have a different genetic potential for height.
In polygenic traits (remember, most traits are polygenic), several genes mutually have a say in the ultimate phenotype that is produced.




No comments:
Post a Comment