Biology Index
Where are we going with this? The information on this page should increase understanding related to this standard: Identify patterns of inheritance to predict genotype/phenotype and solve punnett square problems.
Article includes ideas, images, and content from Troy Smigielski (2022-01)
Introducing Human Genetics:
Sex-linked Inheritance
(Pleased to meet you! See what I did, there?)
In previous articles, we have looked closely at genetics. In this and a few more that will follow, we will apply those to humans as a specific case of some additional general genetics principles.
- Genetic information is carried by DNA.
- DNA is organized into structures called chromosomes.
- Chromosomes carry information for specific traits in sections called genes.
- Genes having different versions or variations of a trait are alleles.
- Some traits are dominant, some are recessive, some are incompletely dominant, and some are codominant.
- Some traits require information from more than one gene; these are polygenic.
- Humans have 46 chromosomes organized into 23 pairs; one chromosome from each pair comes from the mother and the other one comes from the father.
We should probably repeat some of that, but with pictures!
Humans have 23 pairs of each chromosome. That is to say, there are two each of the 23 different chromosomes that are paired up. That makes 46; 23 come from the mother and 23 come from the father.
In other words, humans have two copies of each chromosome. This makes us diploid organisms.
Wooo, that's a cool picture! What's it called?
Biologists use diagrams called karyotypes to map out chromosomes of an organism. For example, there is not a human.
To create a karyotype, biologists take pictures of cells in stages of mitosis. Then, they organize the chromosomes by size and shape. Something like the following…
So, in other words, it's magic?
A karyotype can give you three major pieces of information:
- How many chromosomes an organism has
- Sex of the organism
- Presence of a chromosomal disorder
How can that bunch of dark splotches tell what the sex of an organism is?
Sex/gender is all about the 23rd pair of chromosomes.
The 23rd chromosome pair of a male is XY.
The 23rd chromosome pair of a female is XX.
To indicate an organism biologically, scientists use the number of chromosomes followed by XX or XY for sex. For example, a human male would be 46,XY and a human female would be 46,XX.
The X and Y chromosomes are called sex chromosomes because they determine sex. In humans, these are found on the 23rd pair.
All chromosomes that do not determine sex are called autosomes. In humans, these are the first 22 pairs. Most genes are located somewhere on an autosome.
Determining The Sex of Offspring
So, genetics… all that… genes… chromosomes… all of this must let us figure things out. For instance,
what is the probability that a baby will be a male? A female?
50/50.
How do we know that?
What two sex chromosomes does mom have? X and X…
What two sex chromosomes does dad have? X and Y…
We can find probabilities of the offspring being a certain sex through Punnett squares. Males will either pass on an X or a Y chromosome. Females will pass on an X chromosome no matter what.
Looking at the Punnett square, two of the boxes have XX and two have XY. Hence, half will be mail and half will be female.
Since only the male can pass the Y, then ultimately, the father ultimately the sex of the child.
Somebody should have told Henry XIII… Just saying…
Sex-linked Traits and Disorders
Although most traits are carried on the autosomes, some traits and disorders are carried on the sex chromosomes. These are called sex-linked traits.
Sex-linked traits: traits that are carried on the sex chromosomes.
Other traits that are carried on Chromosomes #1-22 are called autosomal traits. Sex-linked traits and disorders are typically carried on the X chromosome because it is larger and has more space to carry genes. The Y chromosome is shorter and does not carry as many genes. Most sex-linked traits are recessive.
So… Think… If the recessive trait is on the X chromosome, since the male has only one of them, it will be expressed; there's no "competing trait" on the Y chromosome. In the female, there is a chance that the other X chromosome will carry the dominant trait, so the recessive trait won't be expressed.
Let's look at this idea with another image…
In the image above, the Y chromosome does not have the genes labeled A, B, and C.
(This is an example and does not necessarily model real genes exactly.) Therefore, whatever traits are controlled by those genes will come through in the child. The Y chromosome has no alternative to offer; the gene of the X chromosome will be expressed.
Example: Color Blindness
Many people can think of someone who is color blind. If you can, are they male or female? You probably answered that they were male. Why?
Sex-linked traits and disorders are more common in males. Why is that?

Since men only have one X chromosome, whatever is on it will be expressed. Since women have two X chromosomes, they get “two chances”.
Example: Hemophilia
Hemophilia is a recessive, sex-linked disorder. Both males and females can have it.
In the image to the right, the father has the recessive trait on the X chromosome. He has no allele for that trait on the Y chromosome. The female has one X with, and one X without the trait.
Therefore, the presence of hemophilia is determined by which of th X chromosomes the female passes.
The four possible outcomes are shown in among the daughters and sons.
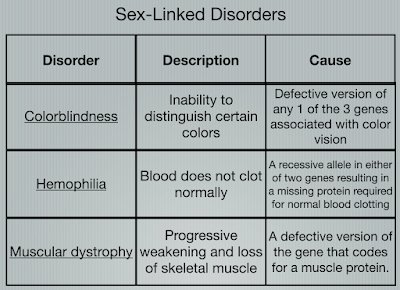
There are numerous sex-linked disorders that come from X chromosome. Thus, these disorders can only be passed on from the mother. Three of them are shown in the image below.
Some Examples of Sex-linked Traits:
- Red-green colorblindness
- Male Pattern Baldness
- Hemophilia
- Duchenne Muscular Dystrophy
Predicting The Inheritance of Sex-linked Traits
We can do Punnett squares to determine the likelihood that offspring will inherit a sex-linked trait.
Below, the gene H is indicated as being part of the X chromosome. The H is the dominant, "unaffected" trait and the h is the recessive "affected" trait.
If we were to cross a normal male with a colorblind female, the results would be predictable using a Punnett square. It would show both the chances for sons and daughters to have the condition.
This is the Punnett square for the above cross:
Note that the female carries the recessive c gene for color blindness on each of the X chromosomes. The male does not have gene for color blindness at all on the Y chromosome and has the dominant gene (not color blind) on his X chromosome.
Hence, the daughters will always get the "not color blind" X chromosome from the father (and the recessive c "color blind" gene from the mother. So, 0% of the daughters will be color blind.
The sons will get the Y chromosome from the father which does not have a gene for color blindness at all. Thus, the Xc chromosome from the mother (she has 2) will always be expressed. Therefore, 100% of the sons will be color blind.
Origins of Sex-linked Trait Understanding
The classic example of X-linked inheritance is eye color of fruit flies (
Drosophila melanogaster).
Thomas Hunt Morgan was working with fruit flies when he noticed that most of the flies that had white eyes were males. (Fruit flies can have either red or white eyes.)
This told him that eye color must be carried on the X chromosome because one is much more common in males. This also told him that red eyes are dominant to white eyes.
Red eyes are the wild type which means they are normal and more common. White eyes are the mutant type which means they are abnormal and less common.
Using Punnett squares, different combinations of different crosses can be examined. Where R is the dominant (red) trait and r is the recessive (white) trait, we get this:
Female Key:
XRXR = red eyes
XRXr = red eyes
XrXr = white eyes
Male Key:
XRY = red eyes
XrY = white eyes
Crossing a heterozygous female with a white-eyed male would result in:
50% of females have red eyes.
50% of males have red eyes.
This process can be repeated, of course, for any variation of the cross.
Non-typical Genetic Outcomes
There are occasions when the normal process does not take pace just right. Normally, the process of meiosis results in four haploid gametes.
Sometimes, the separating process does not go as expected. When chromosomes separate incorrectly during Meiosis, it is called non-disjunction.
That might look something like this (instead of the illustration above).
This leads to aneuploidy which is when one of the gametes has the incorrect number of chromosomes.
Aneuploidy can occur in both the autosomes and in the sex chromosomes. There are a numerous kinds of aneuploidy caused by nondisjunction. A few of them are illustrated below:
Monosomy is when the offspring only received one copy of a chromosome.
Trisomy is when the offspring received three copies of a chromosome.
Down syndrome is when an individual received three copies of chromosome #21. This is also known as Trisomy 21.
Klinefelter’s syndrome is when an individual receives XXY chromosomes; this results in the male phenotype with enlarged breasts.
Turner's syndrome is when an individual receives one X chromosome; this results in the female phenotype with underdeveloped breasts and degenerated ovaries.
Interactions Of Chromosomes
The human body only needs one X chromosome. Women have 2 X chromosomes, so they inactivate one of them. The inactivated chromosome is called a Barr body.
The presence or absence of the disorder is determined as soon as you are created. X-inactivation happens after zygote formation, and some scientists think that a type of decision making process may occur at the cellular level.
A cat with different colors is very likely a female. This is because the fur color gene is on the X chromosome. At different places on the cat’s body, different X chromosomes are inactivated. Therefore, there are different colors in different spots.
For a male to have different colors, he would have to have XXY.